Publications
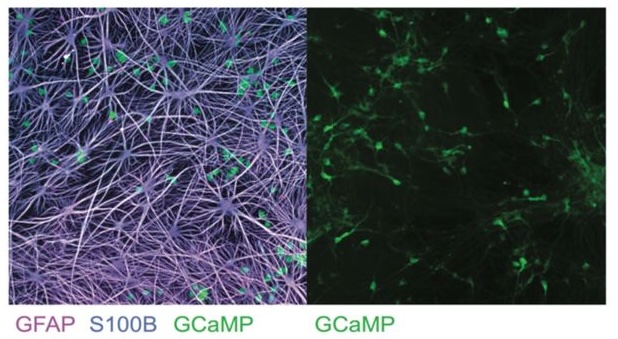
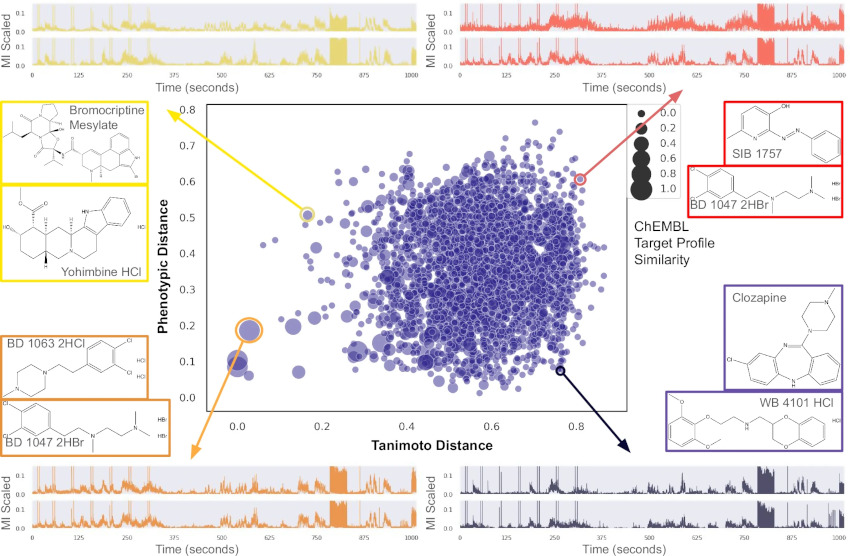
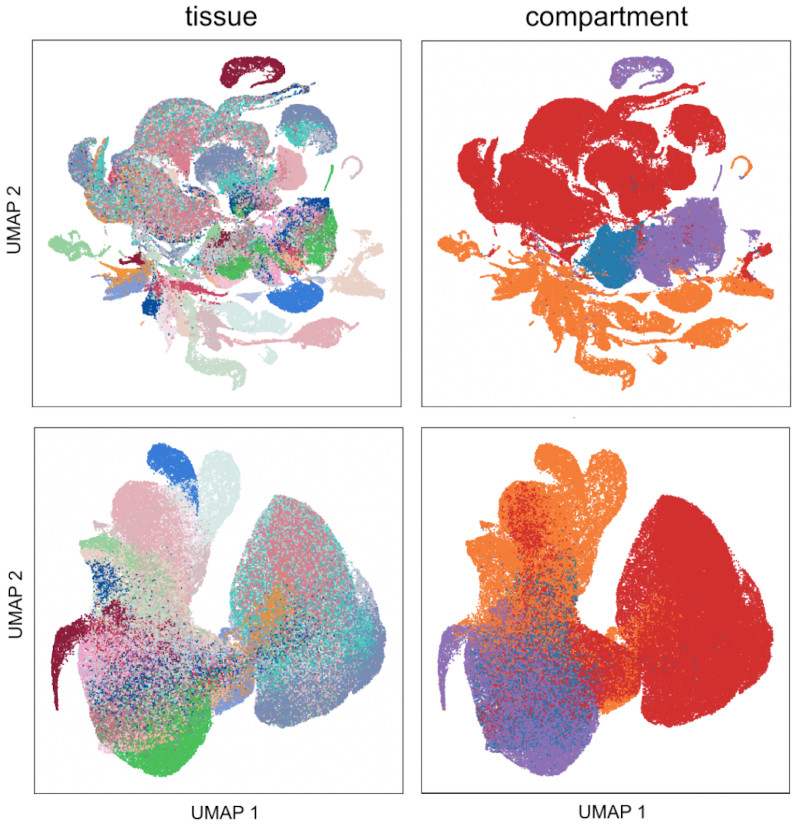
Network-aware self-supervised learning enables high-content phenotypic screening for genetic modifiers of neuronal activity dynamics
bioRxiv. 2025 Feb 05. Grosjean P, Shevade K, Nguyen C, Ancheta S, Mader K, Franco I, Heo SJ, Lewis G, Boggess S, De Domenico A, Ullian E, Shafer S, Litterman A, Przybyla L, Keiser MJ, Ifkovits J, Yala A, Kampmann N.
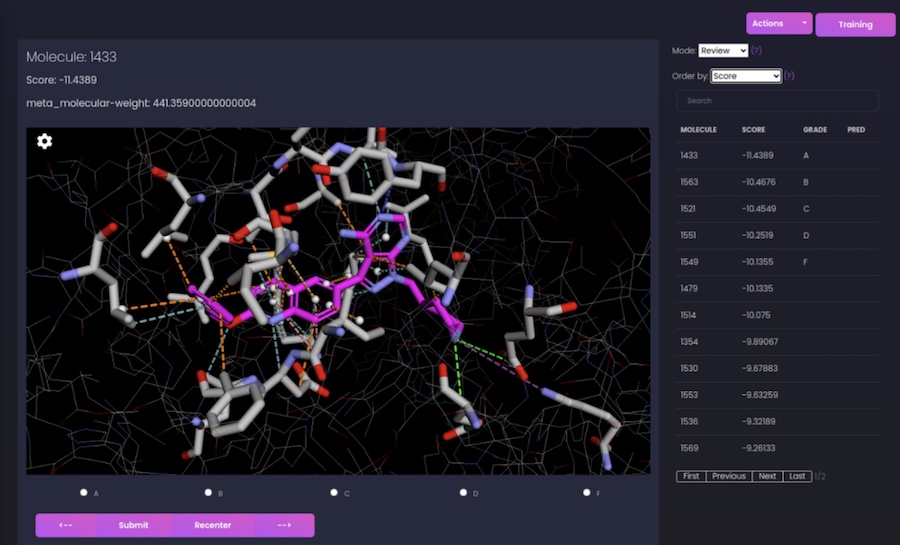
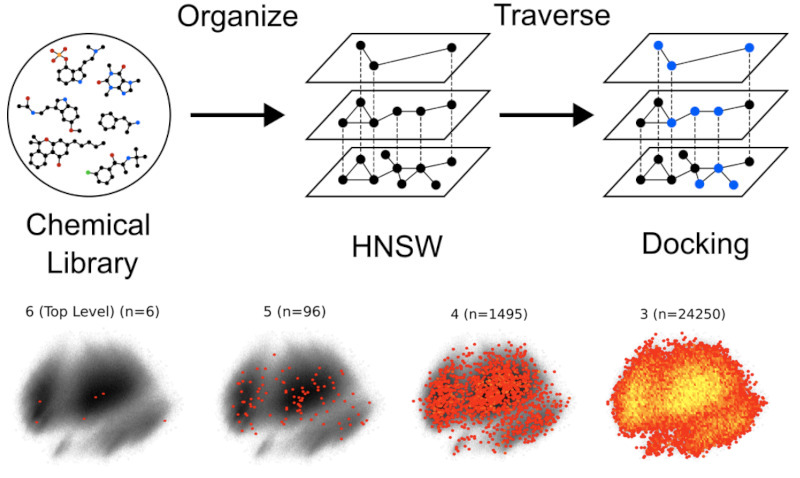
Autoparty: Machine learning-guided visual inspection of molecular docking results
chemRxiv. 2024 Dec 09. Shub L, Korczynska M, Muir DF, Lin F-Y, Mathiowetz AM, Keiser MJ.
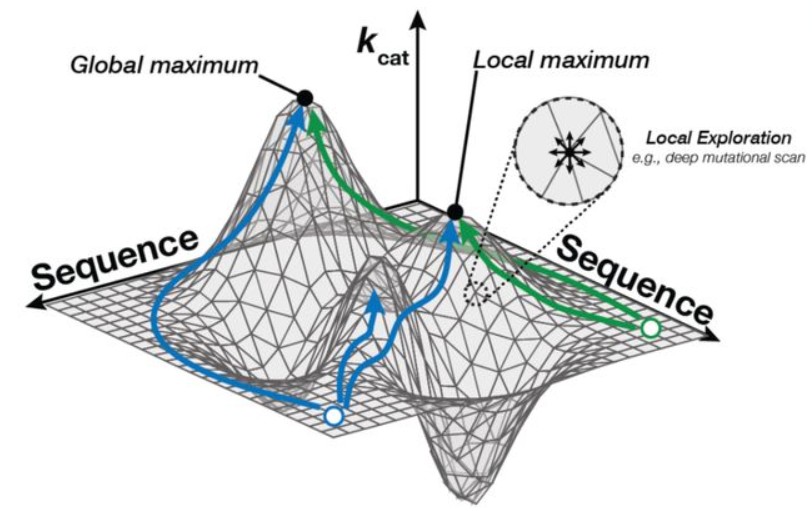
Evolutionary-scale enzymology enables biochemical constant prediction across a multi-peaked catalytic landscape
bioRxiv. 2024 Oct 25. Muir DF, Asper GPR, Notin P, Posner JA, Marks DS, Keiser MJ, Pinney MM.
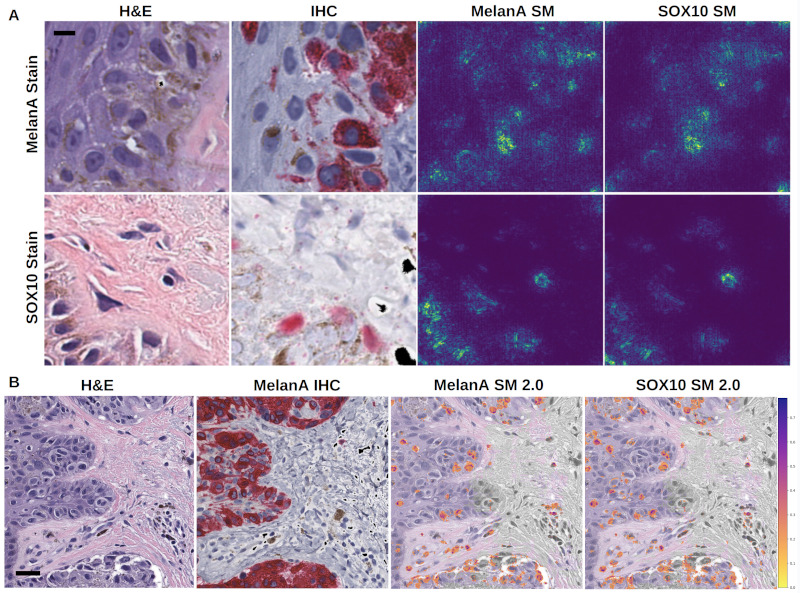
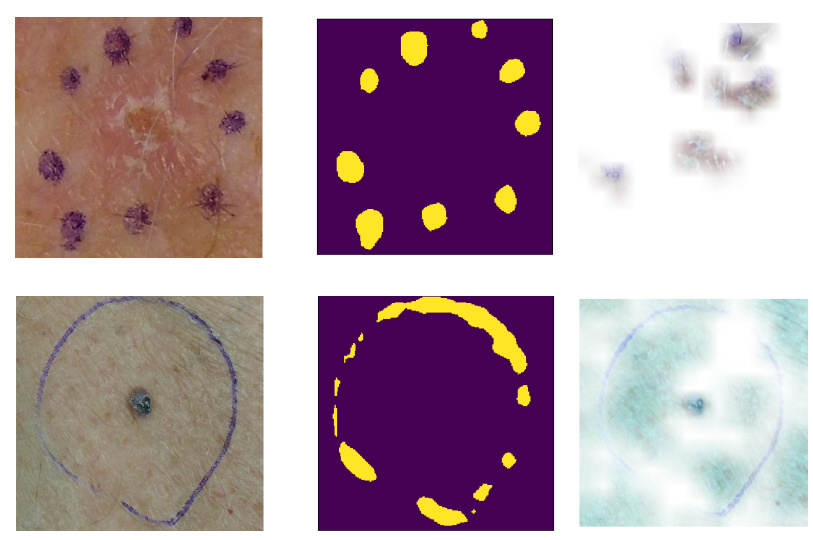
Machine-learning convergent melanocytic morphology despite noisy archival slides
bioRxiv. 2024 Sep 12. Tada M, Gaskins G, Ghandian S, Mew N, Keiser MJ, Keiser ES.
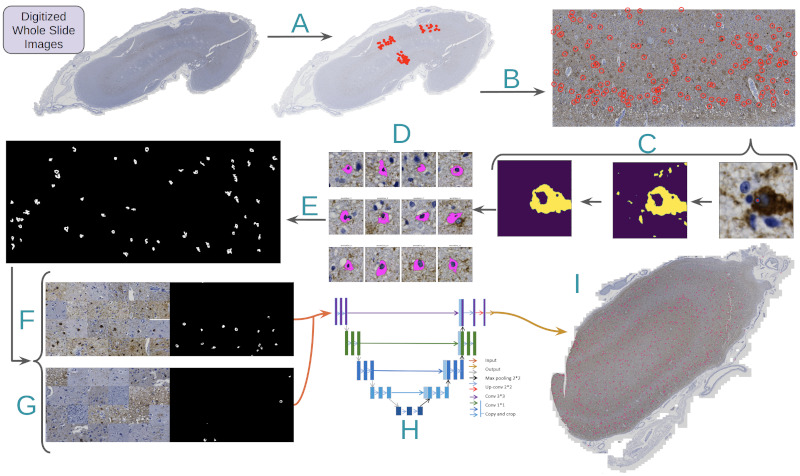
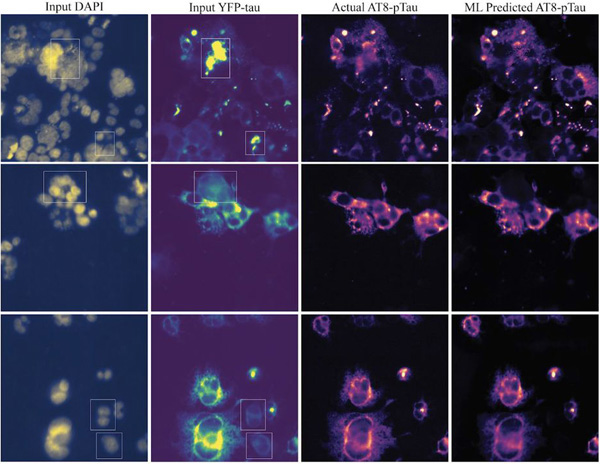
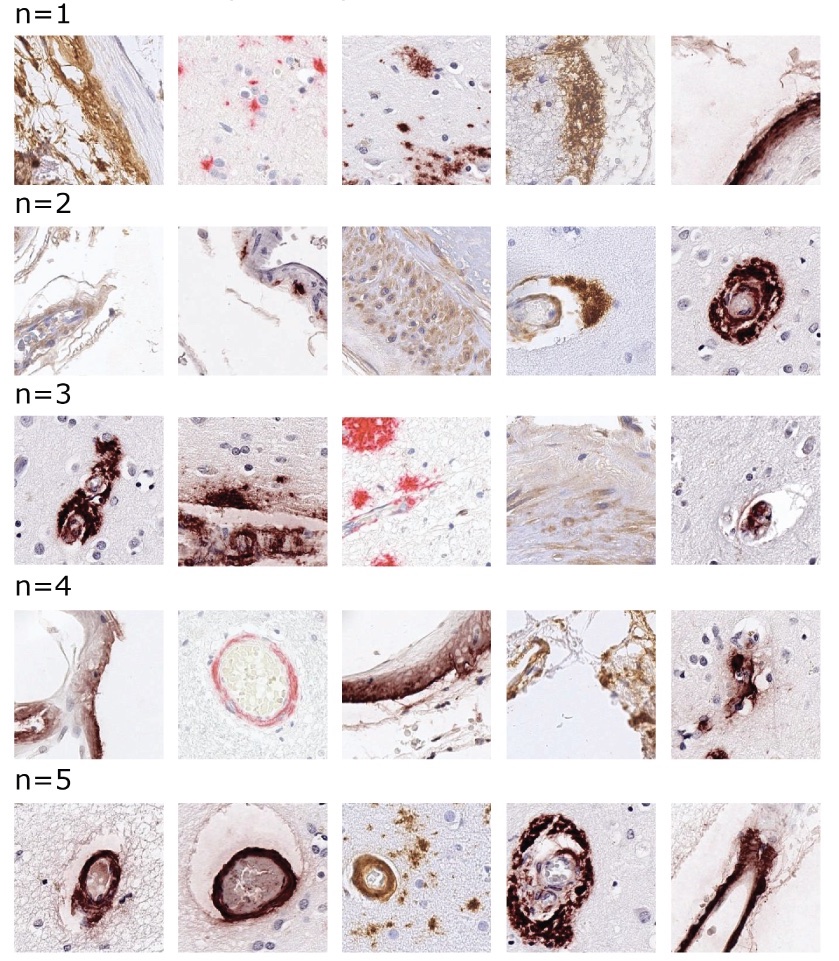
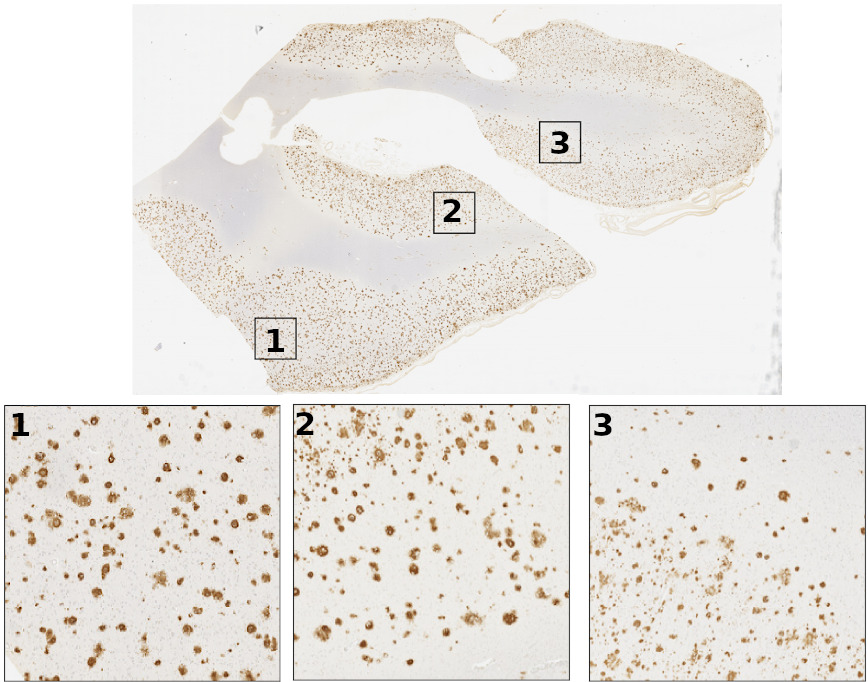
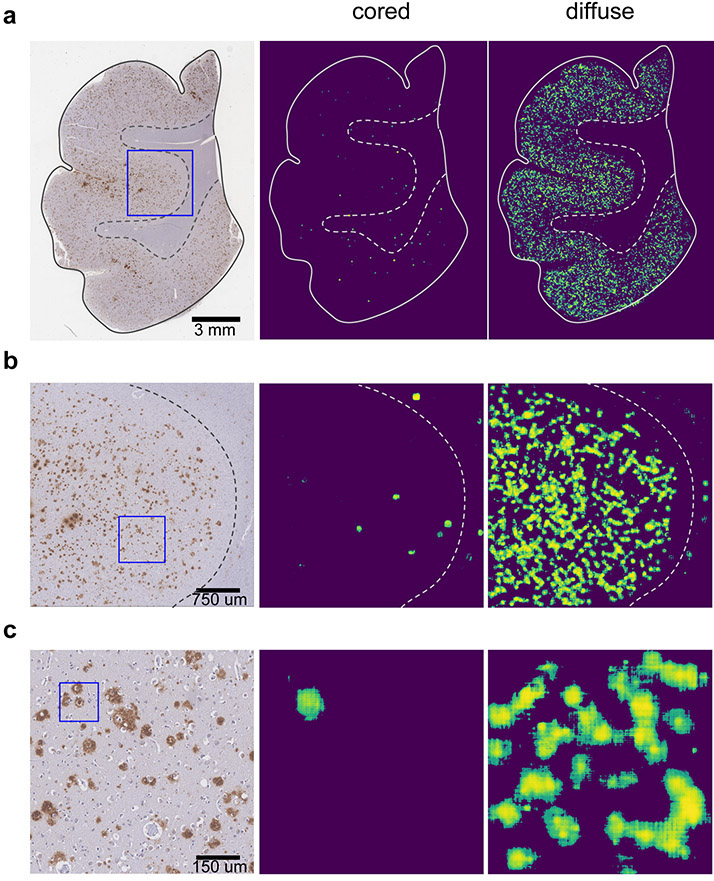
Learning precise segmentation of neurofibrillary tangles from rapid manual point annotations
bioRxiv. 2024 May 15. Ghandian S, Albarghouthi L, Nava K, Sharma SRR, Minaud L, Beckett L, Saito N, DeCarli C, Rissman RA, Teich AF, Jin LW, Dugger BN, Keiser MJ.
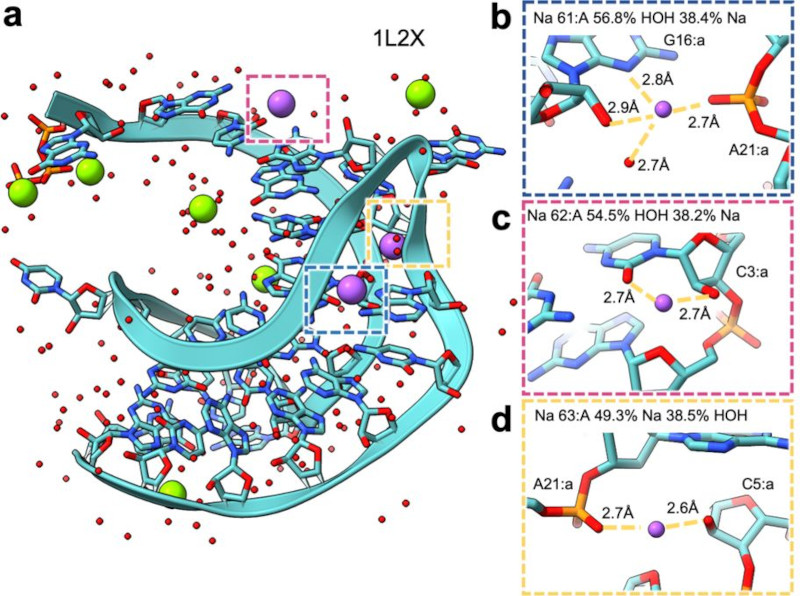
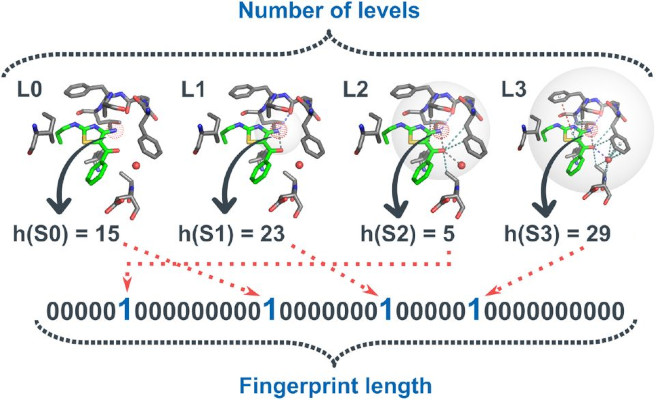
Metric Ion Classification (MIC): A deep learning tool for assigning ions and waters in cryo-EM and x-ray crystallography structures
bioRxiv. 2024 Mar 19. Shub L, Liu W, Skiniotis G, Keiser MJ, Robertson MJ.
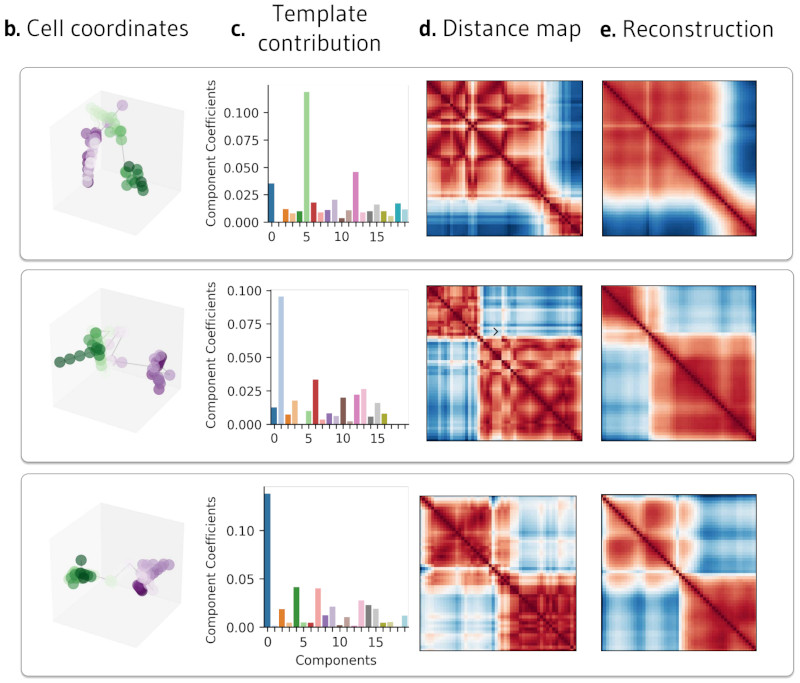
Toward a generalizable machine learning workflow for neurodegenerative disease staging with focus on neurofibrillary tangles
Acta Neuropathol Commun. 2023 Dec 18. Vizcarra JC, Pearce TM, Dugger BN, Keiser MJ, Gearing M, Crary JF, Kiely EJ, Morris M, White B, Glass JD, Farrell K, Gutman DA.
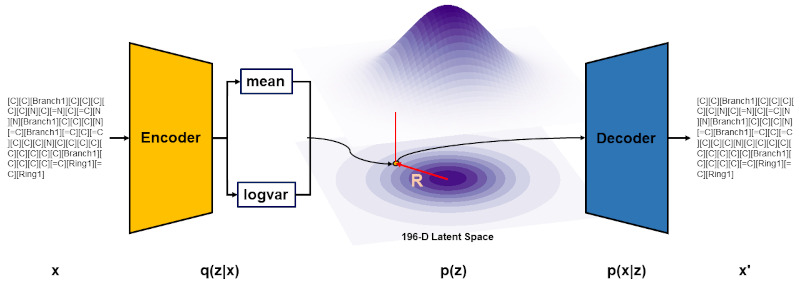
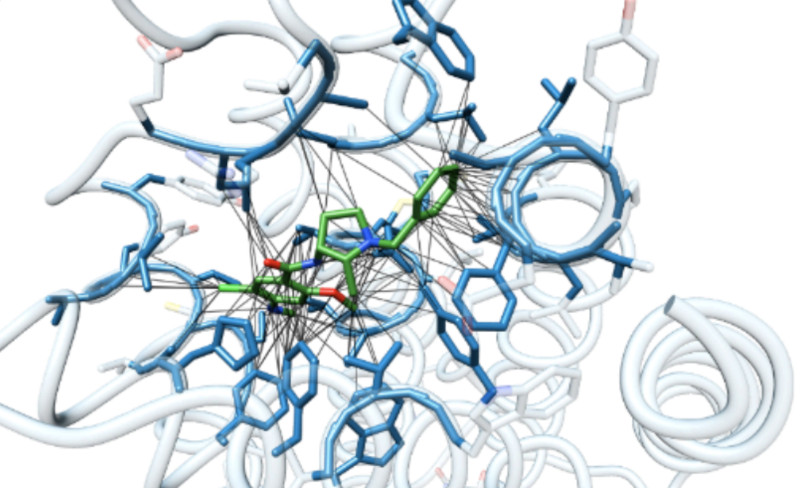
Autoregressive fragment-based diffusion for pocket-aware ligand design
arXiv - NeurIPS GenBio. 2023 Nov 2. Ghorbani M, Gendelev L, Beroza P, Keiser MJ.
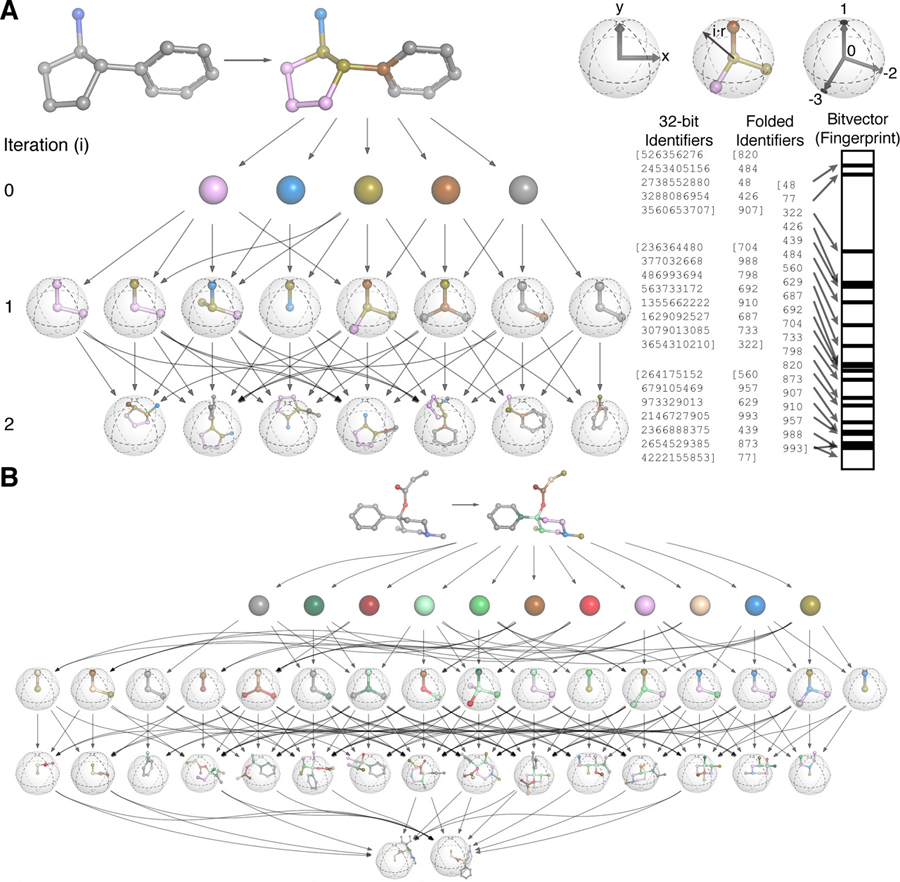
A single-cell gene expression language model
arXiv - NeurIPS LMRL. 2022 Oct 25. Connell W, Khan U, Keiser MJ.
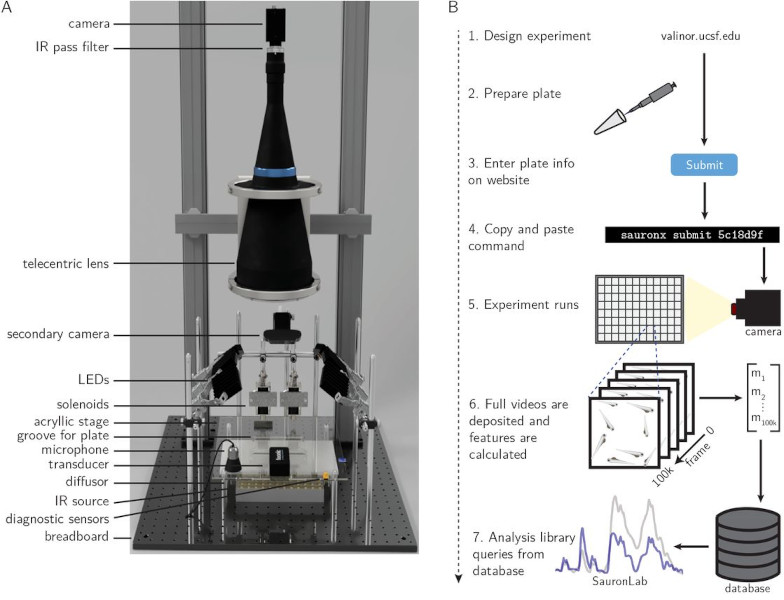
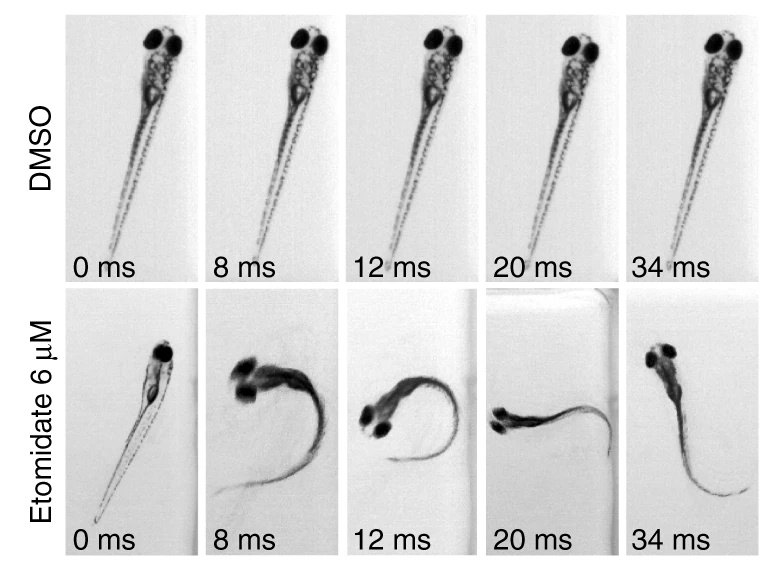
Simultaneous analysis of neuroactive compounds in zebrafish
bioRxiv. 2022 Jun 15. Myers-Turnbull D, Taylor JC, Helsell C, McCarroll MN, Ki CS, Tummino TA, Ravikumar S, Kinser R, Gendelev L, Alexander R, Keiser MJ, Kokel D.
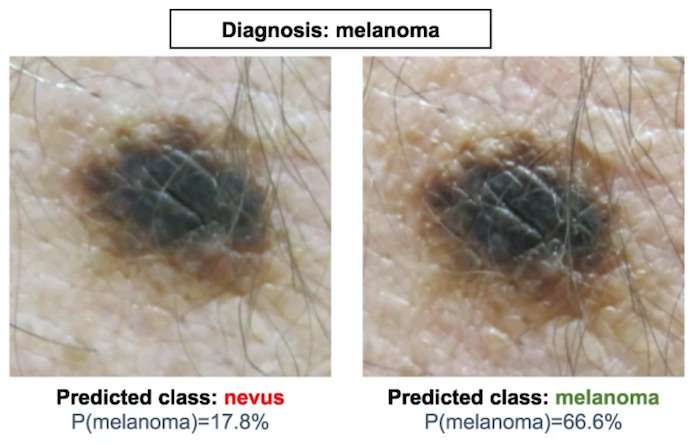
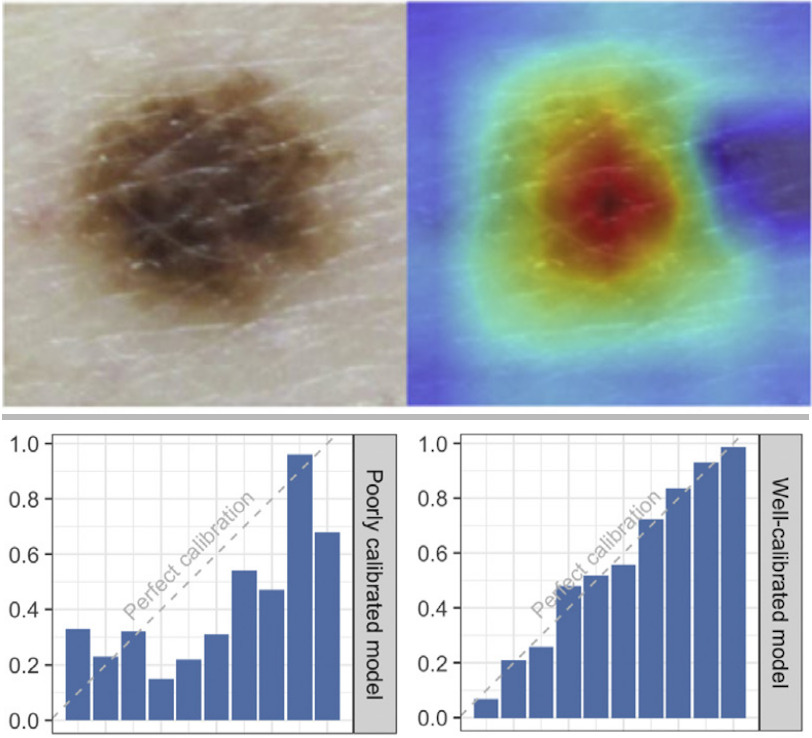
Stress testing reveals gaps in clinic readiness of image-based diagnostic artificial intelligence models
NPJ Digit Med. 2021 Jan 21. Young AT, Fernandez K, Pfau J, Reddy R, Cao NA, von Franque MY, Johal A, Wu BV, Wu RR, Chen JY, Fadadu RP, Vasquez JA, Tam A, Keiser MJ, Wei ML.
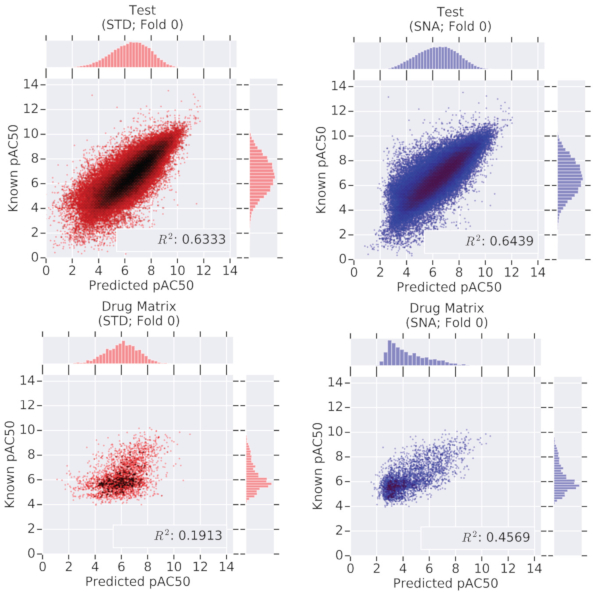
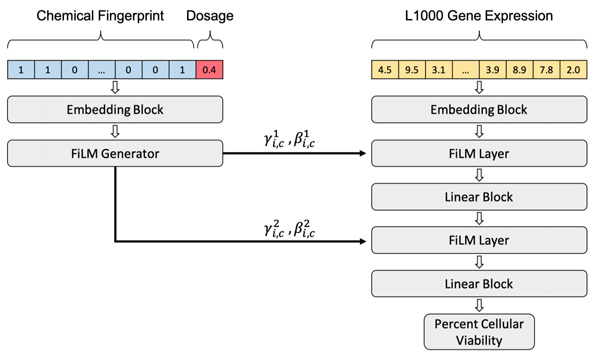
Predicting Cellular Drug Sensitivity using Conditional Modulation of Gene Expression
bioRxiv - NeurIPS LMRL. 2020 Dec 11. Connell W, Keiser MJ.
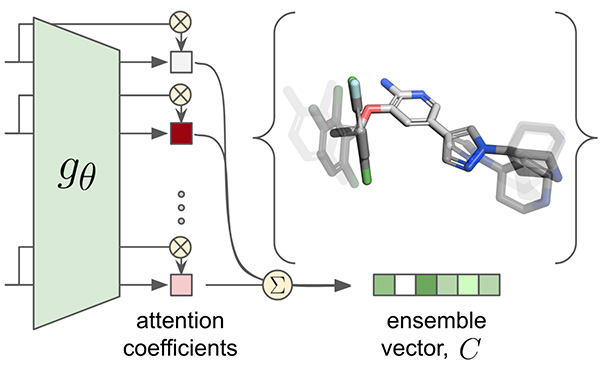
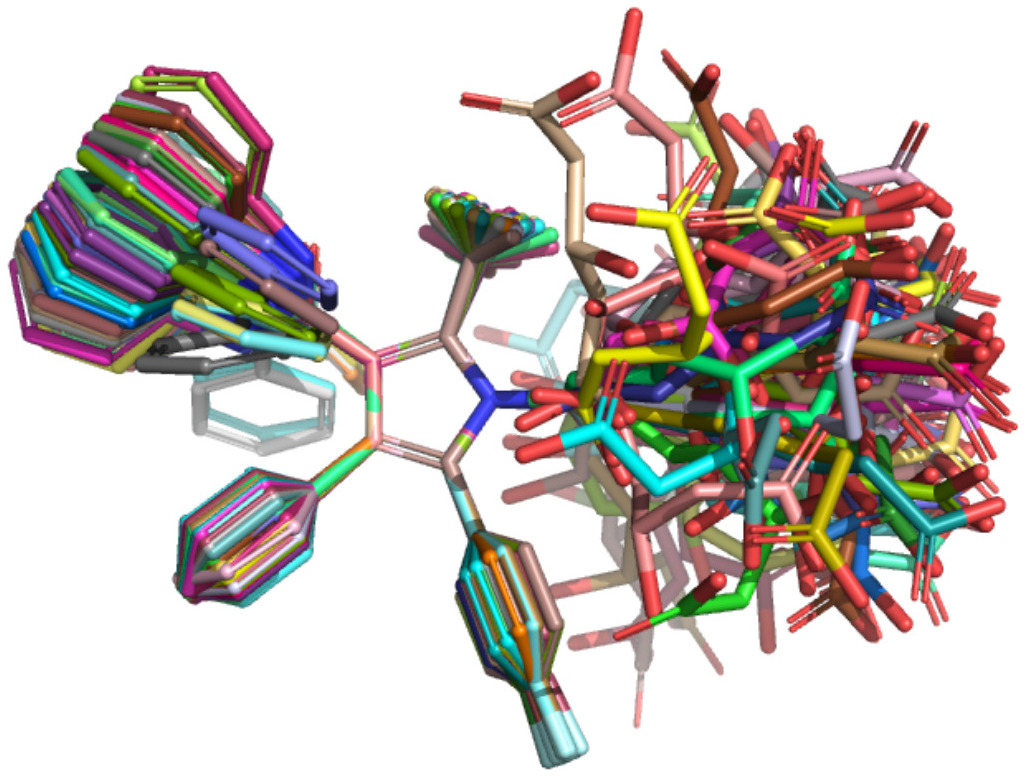
Attention-Based Learning on Molecular Ensembles
arXiv - NeurIPS ML4Molecules. 2020 Nov 25. Chuang KV, Keiser MJ.
Learning Molecular Representations for Medicinal Chemistry
J Med Chem. 2020 Aug 27. Chuang KV, Gunsalus LM, Keiser MJ.
Artificial Intelligence in Dermatology: A Primer
J Invest Dermatol. 2020 Aug. Young AT, Xiong M, Pfau J, Keiser MJ, Wei ML.
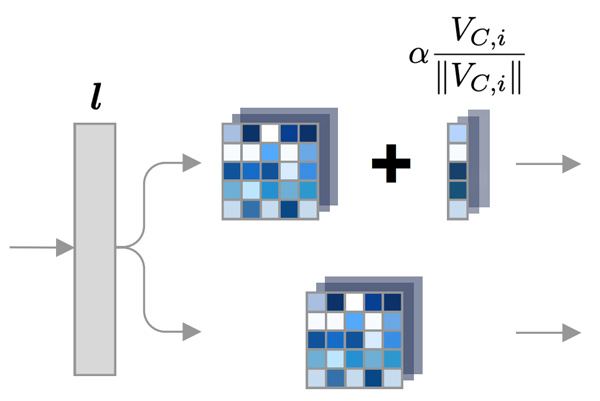
Robust Semantic Interpretability: Revisiting Concept Activation Vectors
arXiv - ICML - WHI. 2020 Jul 17. Pfau J, Young AT, Wei J, Wei ML, Keiser MJ.
Validation of machine learning models to detect amyloid pathologies across institutions
Acta Neuropathol Commun. 2020 Apr 28. Vizcarra JC, Gearing M, Keiser MJ, Glass JD, Dugger BN, Gutman DA.
Global Saliency: Aggregating Saliency Maps to Assess Dataset Artefact Bias
arXiv - NeurIPS ML4H. 2019 Oct 16. Pfau J, Young AT, Wei ML, Keiser MJ.
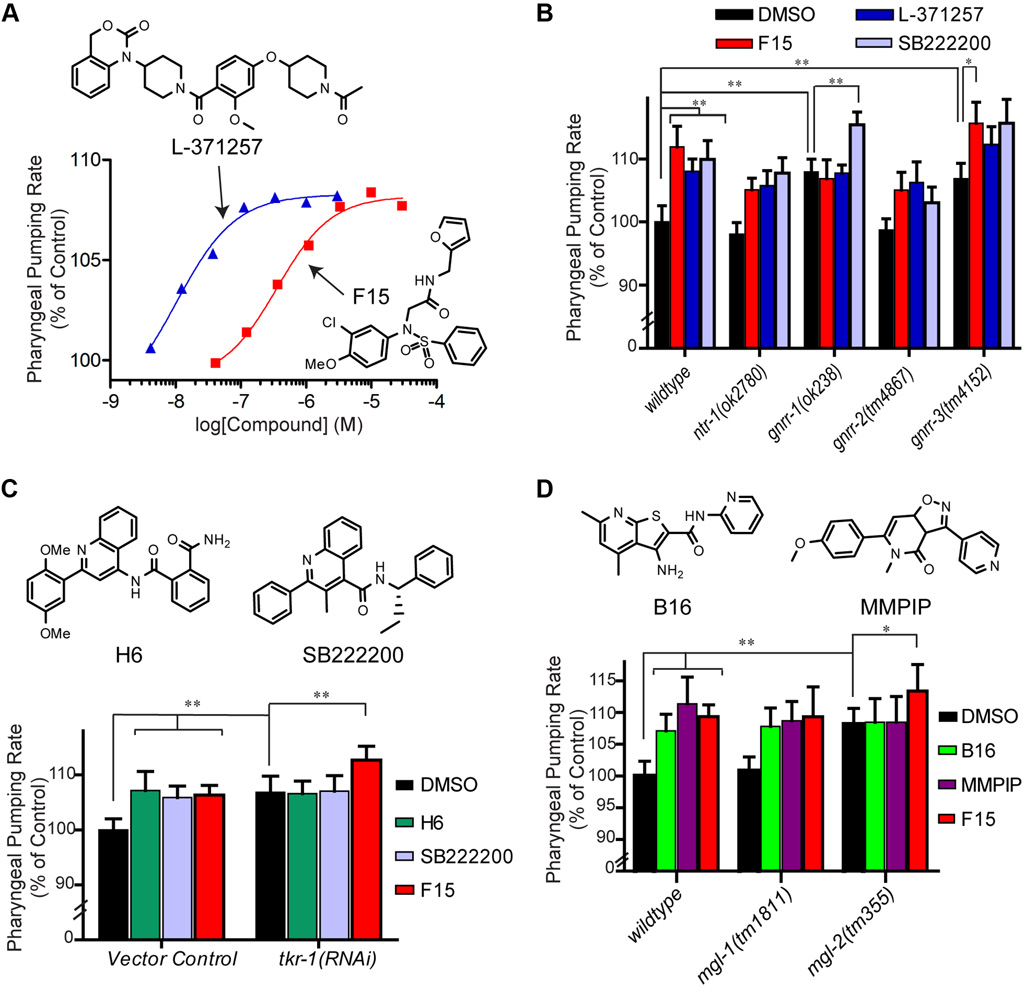
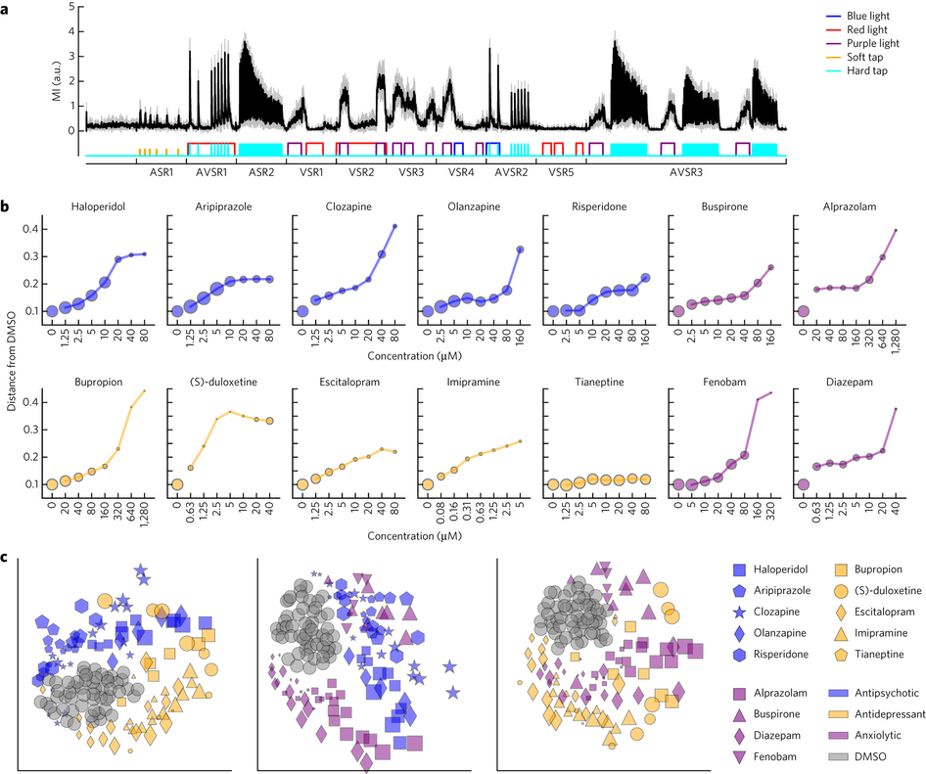
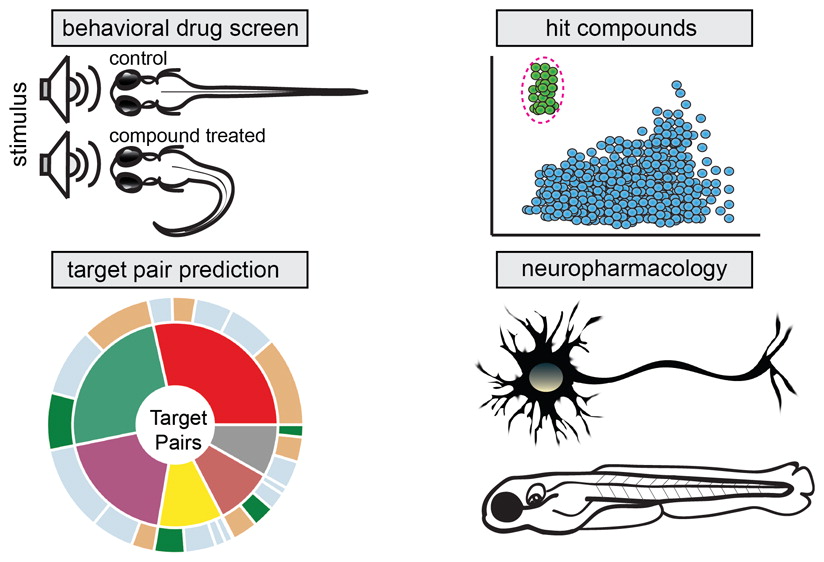
Zebrafish behavioural profiling identifies GABA and serotonin receptor ligands related to sedation and paradoxical excitation
Nat Commun. 2019 Sep 9. McCarroll MN, Gendelev L, Kinser R, Taylor J, Bruni G, Myers-Turnbull D, Helsell C, Carbajal A, Rinaldi C, Kang HJ, Gong JH, Sello JK, Tomita S, Peterson RT, Keiser MJ, Kokel D.
Comment on "Predicting reaction performance in C-N cross-coupling using machine learning"
Science. 2018 Nov 16. Chuang KV, Keiser MJ.
Adversarial Controls for Scientific Machine Learning
ACS Chem Biol. 2018 Oct 19. Chuang KV, Keiser MJ.
The Psychiatric Cell Map Initiative: A Convergent Systems Biological Approach to Illuminating Key Molecular Pathways in Neuropsychiatric Disorders
Cell. 2018 Jul 26. Willsey AJ, Morris MT, Wang S, Willsey HR, Sun N, Teerikorpi N, Baum TB, Cagney G, Bender KJ, Desai TA, Srivastava D, Davis GW, Doudna J, Chang E, Sohal V, Lowenstein DH, Li H, Agard D, Keiser MJ, Shoichet B, von Zastrow M, Mucke L, Finkbeiner S, Gan L, Sestan N, Ward ME, Huttenhain R, Nowakowski TJ, Bellen HJ, Frank LM, Khokha MK, Lifton RP, Kampmann M, Ideker T, State MW, Krogan NJ.
Predicted Biological Activity of Purchasable Chemical Space
J Chem Inf Model. 2018 Jan 22. Irwin JJ, Gaskins G, Sterling T, Mysinger MM, Keiser MJ.
Evolutionarily Conserved Roles for Blood-Brain Barrier Xenobiotic Transporters in Endogenous Steroid Partitioning and Behavior
Cell Rep. 2017 Oct 31. Hindle SJ, Munji RN, Dolghih E, Gaskins G, Orng S, Ishimoto H, Soung A, DeSalvo M, Kitamoto T, Keiser MJ, Jacobson MP, Daneman R, Bainton RJ.
Zebrafish behavioral profiling identifies multitarget antipsychotic-like compounds
Nat Chem Biol. 2016 Jul. Bruni G, Rennekamp AJ, Velenich A, McCarroll M, Gendelev L, Fertsch E, Taylor J, Lakhani P, Lensen D, Evron T, Lorello PJ, Huang XP, Kolczewski S, Carey G, Caldarone BJ, Prinssen E, Roth BL, Keiser MJ, Peterson RT, Kokel D.
Leveraging Large-scale Behavioral Profiling in Zebrafish to Explore Neuroactive Polypharmacology
ACS Chem Biol. 2016 Apr 15. McCarroll MN, Gendelev L, Keiser MJ, Kokel D.
Polygenic overlap between schizophrenia risk and antipsychotic response: a genomic medicine approach
Lancet Psychiatry. 2016 Apr. Ruderfer DM, Charney AW, Readhead B, Kidd BA, Kahler AK, Kenny PJ, Keiser MJ, Moran JL, Hultman CM, Scott SA, Sullivan PF, Purcell SM, Dudley JT, Sklar P.
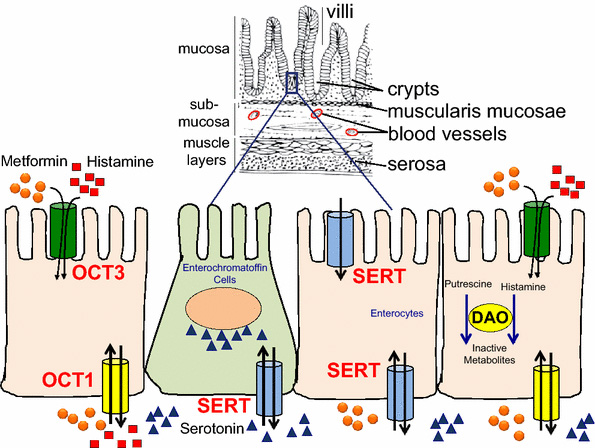
Prediction and validation of enzyme and transporter off-targets for metformin
J Pharmacokinet Pharmacodyn. 2015 Oct. Yee SW, Lin L, Merski M, Keiser MJ, Gupta A, Zhang Y, Chien HC, Shoichet BK, Giacomini KM.
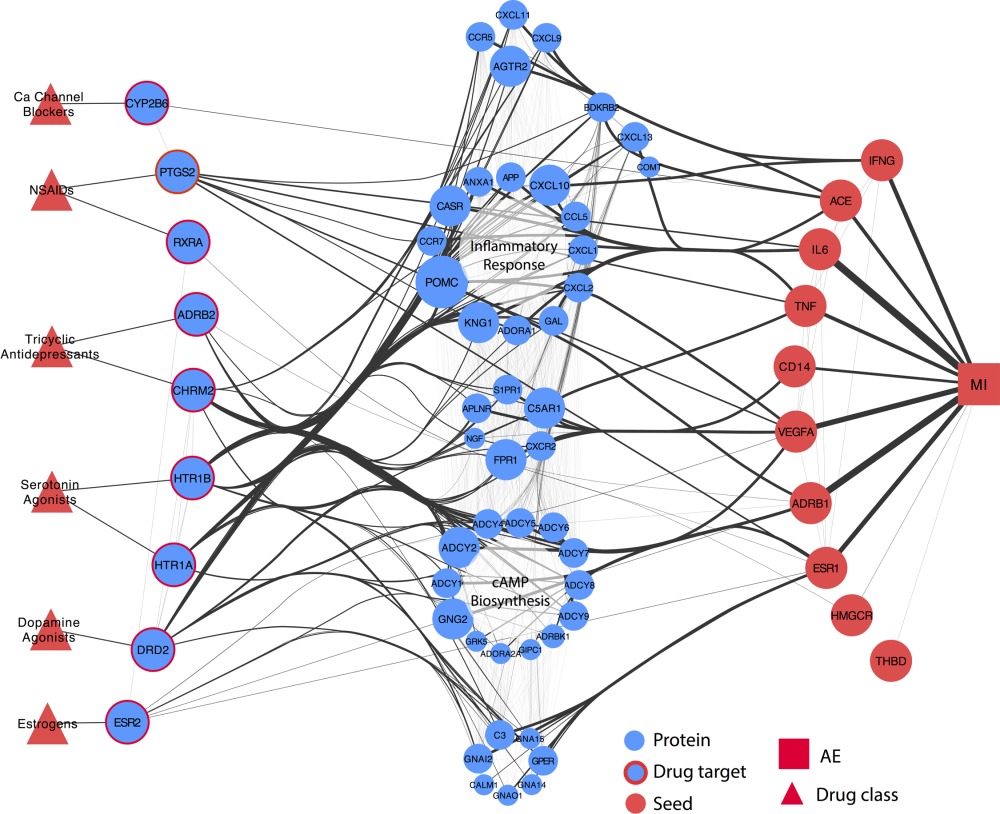
Systems pharmacology augments drug safety surveillance
Clin Pharmacol Ther. 2015 Feb. Lorberbaum T, Nasir M, Keiser MJ, Vilar S, Hripcsak G, Tatonetti NP.
In silico molecular comparisons of C. elegans and mammalian pharmacology identify distinct targets that regulate feeding
PLoS Biol. 2013 Nov. Lemieux GA, Keiser MJ, Sassano MF, Laggner C, Mayer F, Bainton RJ, Werb Z, Roth BL, Shoichet BK, Ashrafi K.
Large-scale prediction and testing of drug activity on side-effect targets
Nature. 2012 Jun 10. Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, Lavan P, Weber E, Doak AK, Cote S, Shoichet BK, Urban L.
Chemical informatics and target identification in a zebrafish phenotypic screen
Nat Chem Biol. 2011 Dec 18. Laggner C, Kokel D, Setola V, Tolia A, Lin H, Irwin JJ, Keiser MJ, Cheung CY, Minor DL Jr, Roth BL, Peterson RT, Shoichet BK.
The presynaptic component of the serotonergic system is required for clozapine's efficacy
Neuropsychopharmacology. 2011 Feb. Yadav PN, Abbas AI, Farrell MS, Setola V, Sciaky N, Huang XP, Kroeze WK, Crawford LK, Piel DA, Keiser MJ, Irwin JJ, Shoichet BK, Deneris ES, Gingrich J, Beck SG, Roth BL.
Complementarity between a docking and a high-throughput screen in discovering new cruzain inhibitors
J Med Chem. 2010 Jul 8. Ferreira RS, Simeonov A, Jadhav A, Eidam O, Mott BT, Keiser MJ, McKerrow JH, Maloney DJ, Irwin JJ, Shoichet BK.
Prediction and evaluation of protein farnesyltransferase inhibition by commercial drugs
J Med Chem. 2010 Mar 25. DeGraw AJ, Keiser MJ, Ochocki JD, Shoichet BK, Distefano MD.
A pilot study of the pharmacodynamic impact of SSRI drug selection and beta-1 receptor genotype (ADRB1) on cardiac vital signs in depressed patients: a novel pharmacogenetic approach
Psychopharmacol Bull. 2010. Thomas KL, Ellingrod VL, Bishop JR, Keiser MJ.
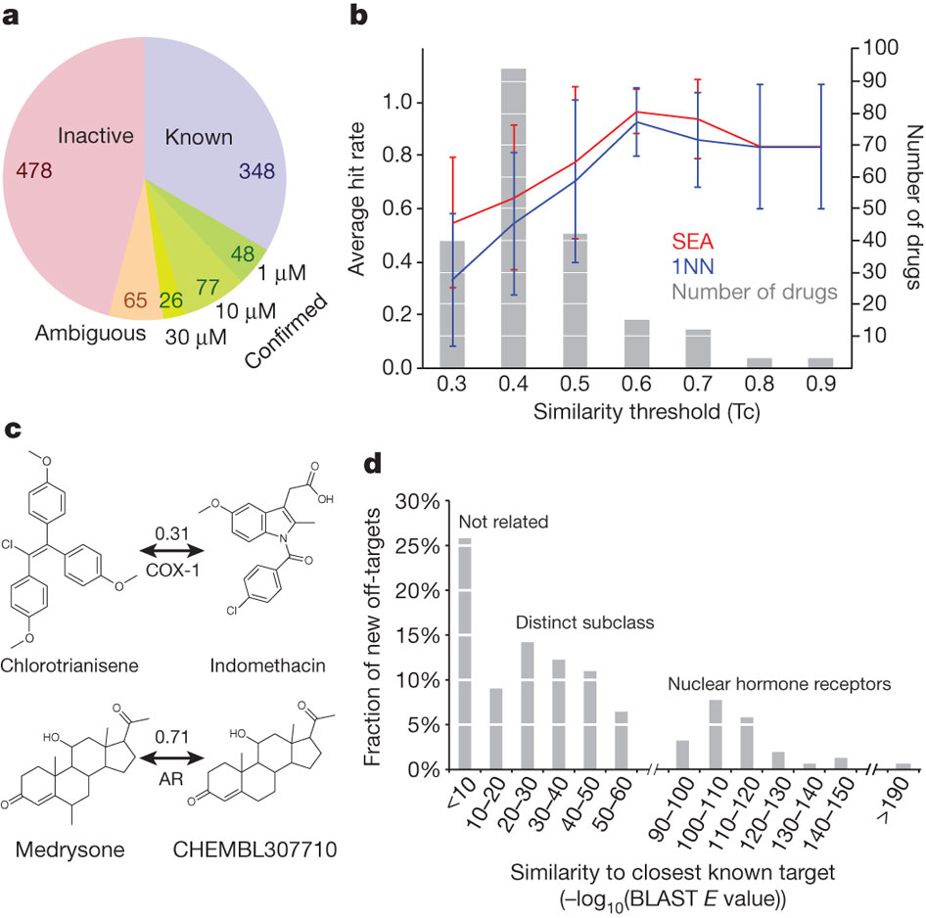
Predicting new molecular targets for known drugs
Nature. 2009 Nov 12. Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, Whaley R, Glennon RA, Hert J, Thomas KL, Edwards DD, Shoichet BK, Roth BL.
A mapping of drug space from the viewpoint of small molecule metabolism
PLoS Comput Biol. 2009 Aug. Adams JC, Keiser MJ, Basuino L, Chambers HF, Lee DS, Wiest OG, Babbitt PC.
Quantifying biogenic bias in screening libraries
Nat Chem Biol. 2009 Jul. Hert J, Irwin JJ, Laggner C, Keiser MJ, Shoichet BK.
Off-target networks derived from ligand set similarity
Methods Mol Biol. 2009. Keiser MJ, Hert J.
Quantifying the relationships among drug classes
J Chem Inf Model. 2008 Apr. Hert J, Keiser MJ, Irwin JJ, Oprea TI, Shoichet BK.
Relating protein pharmacology by ligand chemistry
Nat Biotechnol. 2007 Feb. Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK.


00553-2.jpg)